[演化] 螢光素是怎麼演化出來的? 20191007 PNAS
- Plasmodesmata

- Oct 7, 2019
- 3 min read
Genetically encodable bioluminescent system from fungi
Alexey A. Kotlobay, Karen S. Sarkisyan, Yuliana A. Mokrushina, Marina Marcet-Houbene, Ekaterina O. Serebrovskaya, Nadezhda M. Markina, Louisa Gonzalez Somermeyerc, Andrey, Gorokhovatsky, Andrey Vvedensky, Konstantin V. Purtovh, Valentin N. Petushkov, Natalja S. Rodionova, Tatiana V. Chepurny, Liliia I. Fakhranurova, Elena B. Gugly, Rustam Ziganshin, Aleksandra S. Tsarkova, Zinaida M. Kaskova, Victoria Shender, Maxim Abakumov, Tatiana O. Abakumov, Inna S. Povolotskay, Fedor M. Eroshkin, Andrey G. Zaraisky, Alexander S. Mishin, Sergey V. Dolgov, Tatiana Y. Mitiouchkina, Eugene P. Kopantzev, Hans E. Waldenmaier, Anderson G. Oliveira, Yuichi Oba, Ekaterina Barsova, Ekaterina A. Bogdanova, Toni Gabaldón, Cassius V. Stevani,
Sergey Lukyanov, Ivan V. Smirnov, Josef I. Gitelson, Fyodor A. Kondrashov, and Ilia V. Yampolsky
Abstract
Bioluminescence is found across the entire tree of life, conferring a spectacular set of visually oriented functions from attracting mates to scaring off predators. Half a dozen different luciferins, molecules that emit light when enzymatically oxidized, are known. However, just one biochemical pathway for luciferin biosynthesis has been described in full, which is found only in bacteria. Here, we report identification of the fungal luciferase and three other key enzymes that together form the biosynthetic cycle of the fungal luciferin from caffeic acid, a simple and widespread metabolite. Introduction of the identified genes into the genome of the yeast Pichia pastoris along with caffeic acid biosynthesis genes resulted in a strain that is autoluminescent in standard media. We analyzed evolution of the enzymes of the luciferin biosynthesis cycle and found that fungal bioluminescence emerged through a series of events that included two independent gene duplications. The retention of the duplicated enzymes of the luciferin pathway in nonluminescent fungi shows that the gene duplication was followed by functional sequence divergence of enzymes of at least one gene in the biosynthetic pathway and suggests that the evolution of fungal bioluminescence proceeded through several closely related stepping stone nonluminescent biochemical reactions with adaptive roles. The availability of a complete eukaryotic luciferin biosynthesis pathway provides several applications in biomedicine and bioengineering.
從真菌中發掘,可利用基因體方式編碼的冷光系統
摘要
生物螢光,存在在整個生命演化的各個分支中,提供了特殊視覺性的功能,從吸引異性交配到嚇走掠食者。冷光素(luciferins)經由酵素氧化後,會釋放出光,而我們已知的冷光素已經有約半打。然而,只有一個在細菌中合成冷光速的生化路徑曾經被報導過。在此,我們鑑定出真菌中可將咖啡酸 - 一個簡單卻又廣泛存在的代謝物質,轉化成螢光素的螢光酵素,以及其他三個參與在路徑中的主要酵素。當我們同時將鑑識出來的基因群,以及咖啡酸生合成的基因,轉殖到酵母菌的基因體後,在一般的培養基上,這個轉殖的酵母菌,即可自發螢光。我們研究了這些螢光速生成酵素的演化發現,真菌的生物螢光的出現是經由包含了兩個獨立的基因複製事件的一系列演化步驟所形成。在無螢光的真菌中,複製基因群被保存下來的現象顯示,這個基因複製的發生是在這些基因們,至少其中一個基因參與在生化合成的路徑中,功能分歧之後才發生。這個現象也暗示著,真菌生物螢光的演化過程,是經過許多與環境適應非常相關的非螢光生物化學反應堆積而成。這個完整的真核生物中螢光素的生化合成路徑,在生物醫學以及生物工程上將可能有很大的應用性。
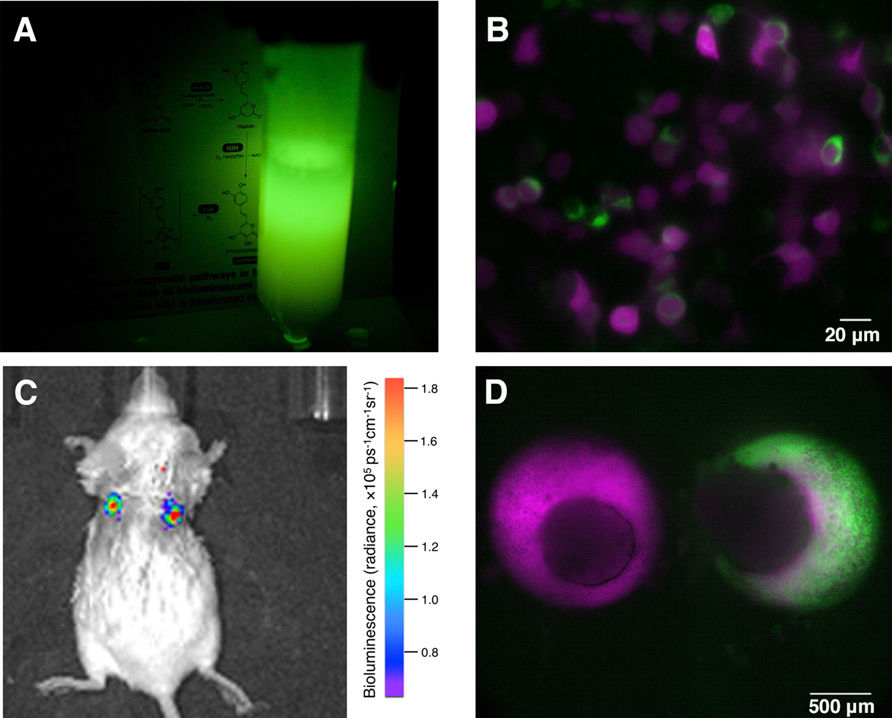
*生物螢光對於很多生物研究有非常重要的角色。這些物質,可以標記一些特定的組織、器官、細胞、乃至於蛋白質,對於追蹤一些特徵,有舉足輕重的地位。而螢光素,是其中一個很早就發現可以發出冷光(化學螢光,不需額外以光源照射即可產生螢光)的化學物質。在使用上,經常是利用一些促進子在特殊細胞表現冷光酵素,當需要測定冷光時,再加入冷光素前驅物來合成冷光素。但是這會造成在生物體中測定有相對的困難,由於角質側或是一些生理特性,導致前驅物不容易到達,那標定的效果就會減低。也因此,大多數的活體標定,目前都還是以綠色螢光蛋白為主體的標定方式。然而螢光蛋白質需要高強度的鐳射照射以激發螢光,對於所標定的細胞以及所希望能觀察的現象,都可能有一定程度的干擾。這篇研究所發現的冷光素合成路徑,可以將所有的基因在同一個細胞、組織或器官中表現,接著這些部位就會自體發出螢光,可以避免許多雷射造成的干擾,也許會是一個不錯的研究工具。




![[植物生化] 在ON/OFF都能提供功能的蛋白質。20191204 SCIENCE](https://static.wixstatic.com/media/a27d24_8cc8e6bd0a064c4a8d7ae6406b9e6aad~mv2.jpg/v1/fill/w_649,h_312,al_c,q_80,enc_avif,quality_auto/a27d24_8cc8e6bd0a064c4a8d7ae6406b9e6aad~mv2.jpg)
![[生物技術] 改造大腸桿菌來固碳,能否為環境變遷帶來新的契機? 20191129 Cell](https://static.wixstatic.com/media/a27d24_c557b52b30d647d5ab23d3ffe5b7bf58~mv2.jpg/v1/fill/w_573,h_570,al_c,q_80,enc_avif,quality_auto/a27d24_c557b52b30d647d5ab23d3ffe5b7bf58~mv2.jpg)
![[植物演化] 離層酸ABA受器的來源。20191128 PNAS](https://static.wixstatic.com/media/a27d24_cf33efcbe4b647819f921d363b98cbf9~mv2.jpg/v1/fill/w_551,h_401,al_c,q_80,enc_avif,quality_auto/a27d24_cf33efcbe4b647819f921d363b98cbf9~mv2.jpg)
Comments